Introduction
It is possible in LinkR to manage multiple data sources (in OMOP format).
These data sources can each be reused in multiple projects.
It is possible to import data from different sources:
- A relational database (DuckDB, PostgreSQL...)
- Parquet files
- CSV files
It will soon be possible to import data in a custom format (data collection) and in FHIR format.
Create a Dataset
To import data, go to the Datasets page from the menu at the top of the screen.
Then click on the Plus (+) icon on the left side of the screen to create a new dataset.
Choose a name. For the example, we will import the MIMIC-III dataset.
For more information about the MIMIC database, go here.
Once the set is created, click on the widget corresponding to this set and go to the Code tab on the right side of the screen.
You will see that R code has been automatically generated.
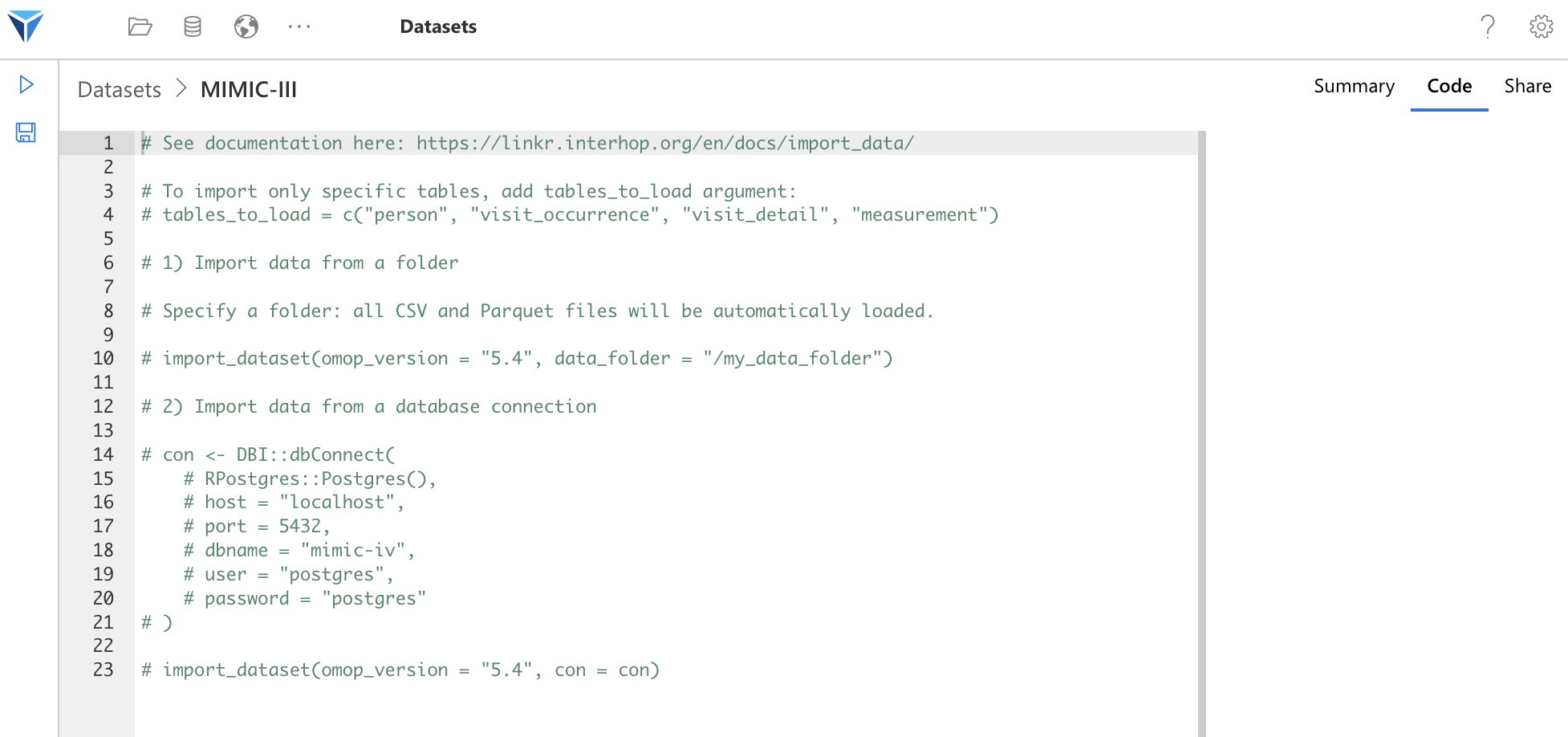
This code gives you two examples of using the import_dataset function, which we will see in detail.
import_dataset Function
To import data into LinkR, we use the import_dataset function.
Here are the function arguments, which we will use depending on the type of data to import (file or database connection):
- omop_version: This is the OMOP version of the data you are going to import (“5.3” or “5.4”)
- data_folder: In the case of importing data from a folder, this is the folder containing the data
- con: In the case of importing data from a database, this is the database connection object
- tables_to_load: By default, all OMOP tables will be loaded from the indicated source. If you only want to load some of these tables, specify here the tables to import. For example
load_tables = c('person', 'visit_occurrence', 'visit_detail')
Database Connection
You can import data from a connection to a database.
First configure the connection object con with the DBI library, then use the import_dataset function.
The con argument will take our con object as value.
# Connection object. We see this in detail below.
con <- DBI::dbConnect(...)
# Function to load data when loading the project
import_dataset(omop_version = "5.4", con = con)
This code will establish a connection to the database when loading a project using this dataset.
Here is an example with a connection to a local PostgreSQL database.
# Connection to local PostgreSQL database
con <- DBI::dbConnect(
RPostgres::Postgres(),
host = "localhost",
port = 5432,
dbname = "mimic-iv-demo",
user = "postgres",
password = "postgres"
)
# Loading data when launching the project
import_dataset(omop_version = "5.4", con = con)
Import Files
You can also import files without going through a database connection.
For this, specify the file location in the data_folder argument.
For example, let’s say my database files are in the /data/mimic-iv-demo/ folder:
/data/mimic-iv-demo/
--- person.parquet
--- visit_occurrence.parquet
--- visit_detail.parquet
--- measurement.parquet
I load them like this.
import_dataset(omop_version = "5.4", data_folder = "/data/mimic-iv-demo/")
Function Execution
Once your script is configured, you can execute the code with the “Execute” button on the left side of the screen (Play icon), or using the CTRL/CMD + SHIFT + ENTER shortcut.
If the data is correctly imported, you will have the row count per table like this.
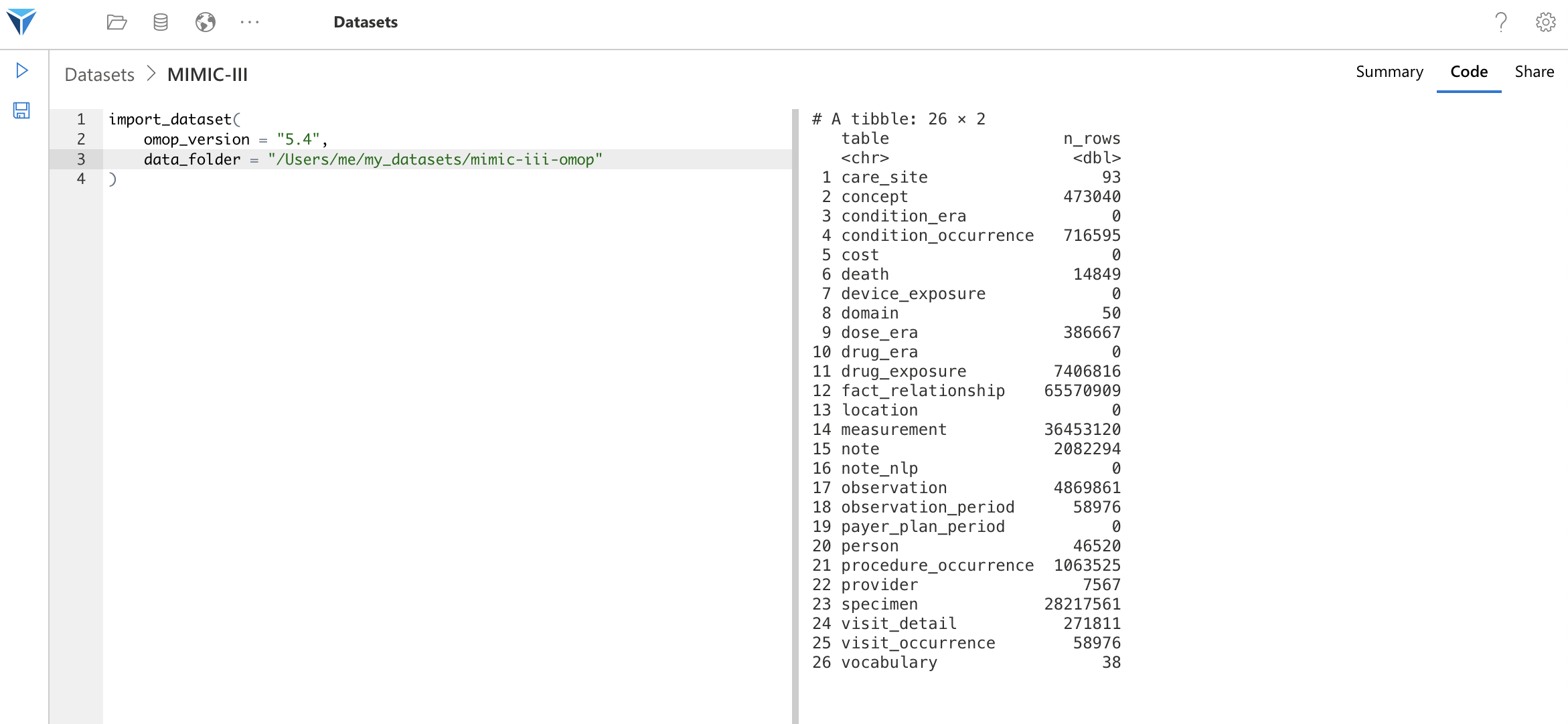
In case of error, the error message will be displayed in this field (on the right side of the screen).
So it’s this script that will execute every time you load a project that uses this dataset.
You can create more complex scripts, which will for example download data from an external source, transform it and save it locally. This is for example what we do with the script to download the MIMIC-IV demo database.
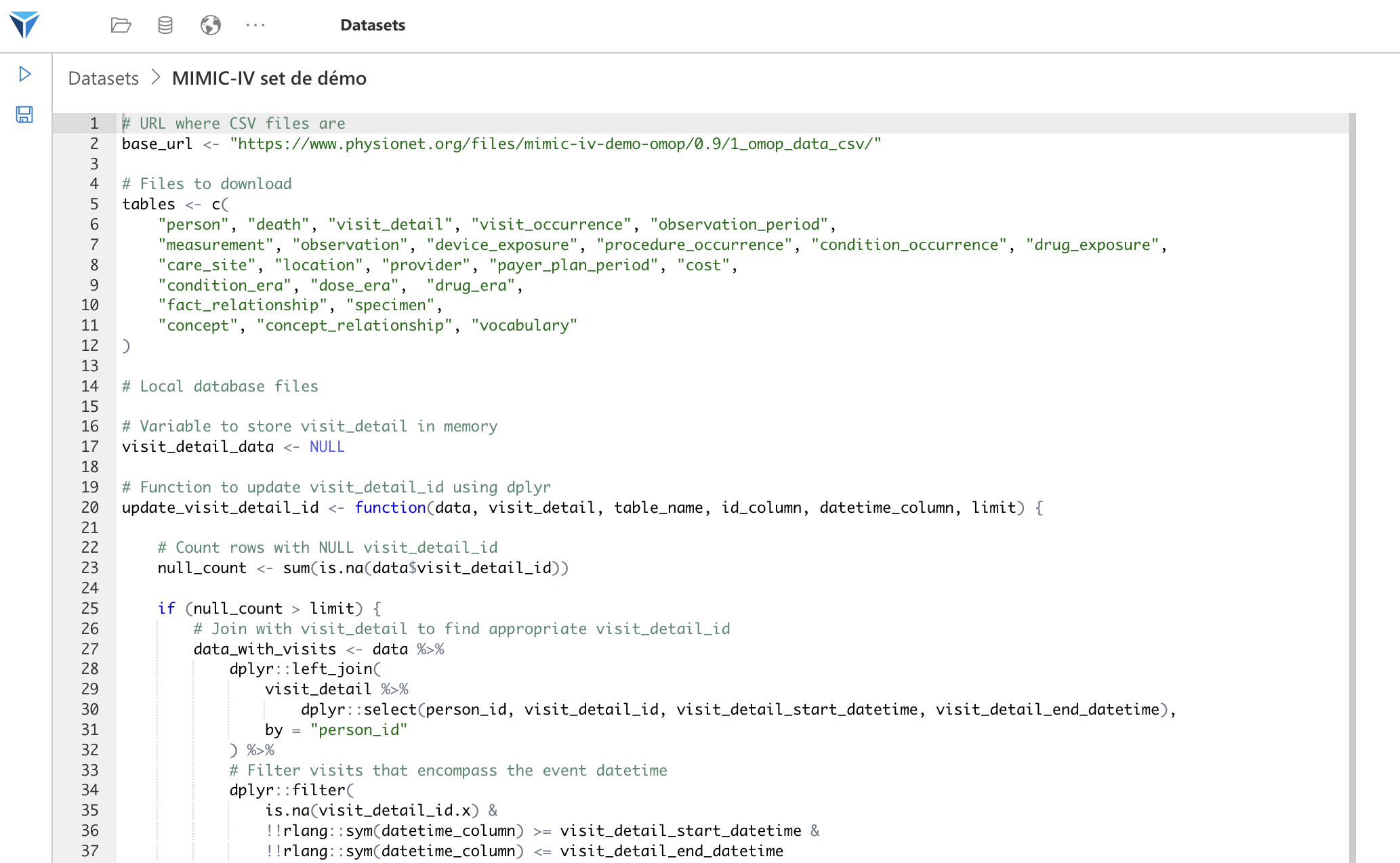
Load Specific Tables
You can choose to import only certain tables from the database, with the tables_to_load argument.
You just need to specify in a character vector the tables to import, like this:
# Loading only person, visit_occurrence, visit_detail and measurement tables
tables <- c("person", "visit_occurrence", "visit_detail", "measurement")
# Adding the tables_to_load argument in import_dataset
import_dataset(omop_version = "5.4", con = con, tables_to_load = tables)
From the Content Catalog
You can also install a dataset from the content library.
This will allow you to download the code to load data (and only the code, we never download data from the content catalog).
Find the tutorial here.
Conclusion




