Introduction
LinkR is organized around projects, in which datasets are loaded, containing data in the OMOP format and using standard vocabularies.
These data can be cleaned and improved using data cleaning scripts.
Within these projects, users can visualize and analyze data using widgets, which are configured plugins.
It is also possible to access a development environment for R and Python via the console.
Datasets
LinkR works with OMOP, an international common data model for healthcare data.
You can import data from various sources: a database, Parquet files, or CSV files.
The same dataset can be used in multiple projects.
Currently, importing data requires writing an R script. A graphical interface will be developed in a future version.
See how to import data.
Vocabularies
The OMOP data model is based on standard vocabularies such as:
- LOINC for lab results
- SNOMED for diagnoses
- RxNorm for medications
- etc.
All these terminologies are available on Athena.
These vocabularies must be imported into LinkR to display the names of the concepts corresponding to concept IDs (the concept_id columns in OMOP tables).
Data cleaning scripts
Data imported in the OMOP format often need to be cleaned using data cleaning scripts.
A common example is weight and height data, which often contain outliers due to how clinical software is designed, such as swapped weight and height fields.
Scripts to exclude such outliers are often created. LinkR facilitates sharing these scripts, which, because of the use of the common OMOP data model, can work across different datasets imported into LinkR.
Other script examples include:
- Calculating scores, such as APACHE-II or the SOFA score
- Calculating urine output by summing different parameters (e.g., urinary catheter, nephrostomy, etc.)
- etc.
Projects
A project is an R and Python environment where data is analyzed.
A project may correspond to a study (e.g., a study on mortality prediction) or to data analysis outside a study, such as creating dashboards (e.g., a dashboard visualizing hospital department activity).
When creating a project, the user selects the data to use from the datasets loaded into the application.
The project will center around two main pages:
-
Individual data page: here, users can recreate the equivalent of a clinical record by creating tabs configured with widgets. For example:
- A “Hemodynamics” tab to configure widgets displaying heart rate, blood pressure, and antihypertensive treatments received by the patient.
- A “Notes” tab to display all textual documents related to the patient (e.g., hospital reports, daily clinical notes).
- An “Infectiology” tab to display all data related to infectiology (e.g., microbiological samples, antibiotics received).
- etc.
-
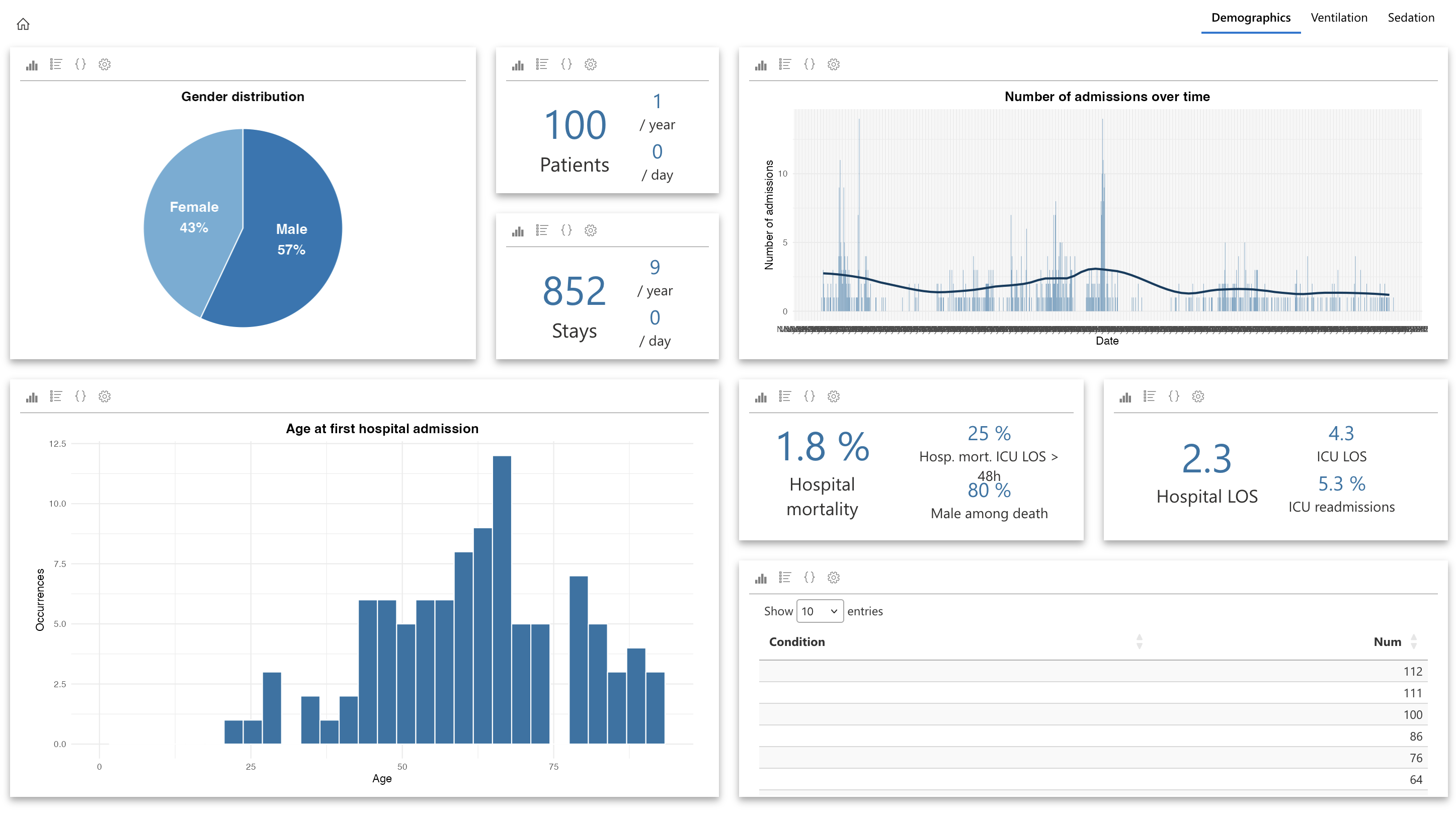
Aggregated data page: here, users can similarly create tabs to configure widgets for group analyses, such as:
- A “Demographics” tab displaying demographic data for a group of patients (e.g., age, sex, length of stay, mortality).
- An “Outlier Data” tab showing distributions of various parameters and excluding outliers.
- A “Survival Analysis” tab with a widget configured for population survival analysis.
- etc.
Using the low-code interface (which combines a code interface and a graphical interface), collaboration between data scientists, statisticians, and clinicians becomes easier.
Plugins
Plugins are blocks of R and Python code that add functionalities to LinkR.
As described earlier, projects are structured with tabs.
These tabs contain widgets, which are plugins applied to data.
For example, if I choose the “Timeline” plugin to be applied to the “Heart Rate” parameter, the resulting widget will be a timeline chart displaying the patient’s heart rate.
There are individual data plugins, which allow recreating a medical record. Examples include:
- Document viewer: displays textual documents (e.g., hospital reports, clinical notes) and filters them (e.g., keyword search, title-based filters).
- Timeline: displays temporal data as a timeline, as described above.
- Datatable: displays data in a tabular format, such as lab results by sampling time.
- etc.
We also have aggregated data plugins for visualizing and analyzing aggregated data, such as:
- ggplot2: a plugin displaying variables using different charts from the ggplot2 library.
- Survival analysis: conducts survival analysis.
- Machine learning: trains and evaluates machine learning models using R or Python libraries.
- etc.
Widgets
Widgets are plugins applied to data.
After creating a tab, you can add multiple widgets to it.
Widgets can be resized and moved around on the page.
Subsets
Within a project, a dataset can be divided into multiple subsets.
A subset is a part of the global dataset, created by applying filters to select patients.
Examples of subsets for the MIMIC database (which contains records for over 50,000 ICU patients, both adult and pediatric) include:
- Patients over 18 years old admitted to the medical ICU for COVID-19.
- Patients with an ICD-10 code for infectious pneumonia treated with Amoxicillin.
- Excluded patients: It may be useful to create a subset with only excluded patients.
- etc.
Currently, creating subsets requires writing R scripts. A graphical interface will be developed in a future version.